Morphological Characterization of Some Cowpea Accessions from Africa
| Received 12 Jan, 2023 |
Accepted 15 Mar, 2023 |
Published 31 Mar, 2023 |
Background and Objective: Cowpea is a pantropical and highly variable crop plant with diverse varieties. The importance of morphological characters in resolving complex differences that coexist among this genus cannot be overemphasized. This study aimed at assessing the qualitative and quantitative characters of ten cowpea accessions collected from ten African countries. Materials and Methods: Cowpea seeds were planted in planting pots with 5 replicates and arranged in an incomplete randomized block design. A total of 37 morphological quantitative and qualitative characters were studied for 12 weeks. Results: There was high variability in the quantitative characters assessed and only TVu-13840 had 100% germination. The TVu-10847 recorded the highest average plant height, mean rachis and petiole length while Tvu-1236 had the highest leaf surface area with no flower pigmentation. Growth and leaf shape varied greatly and the twining tendency was more pronounced in Tvu-10847 and Tvu-11825. All the accessions developed a straight pod shape with the exception of Tvu-14316 which was slightly curved. High variability was observed in raceme position, flower color, seed shape and other distinctive qualitative features. Cluster analysis resolved the characters studied and grouped the accessions into four clusters. Conclusion: The morphological characters of the studied cowpea accessions is an important step in the management of cowpea genetic diversity and also a prerequisite towards selecting desirable traits or improved varieties for breeding purposes.
| Copyright © 2023 Onuminya et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Vigna unguiculata (L.) Walp. commonly known as cowpea is a grain legume belonging to the family Fabaceae. It comprises ten wild perennial subspecies and one annual subspecies (ssp. unguiculata)1,2. It is an herbaceous annual which grows at a temperature of 18-28 throughout all its stages of development3. Cowpea grows well both under favorable and unfavorable growing conditions being able to survive water, temperature and other abiotic stresses4. The dry grain is the most commonly used part of the plant however the fresh or dried leaves5,6, fresh peas and fresh green pods have also been found to be of use in certain cultures. The grain is one of the cheapest sources of proteins which make it extremely valuable to people who cannot afford protein-rich food such as meat and fish in the West and Central Africa, where cowpea is also an important crop7. It is truly a multifunctional crop, with high income generating potential for grain traders and farmers in growing locations.
Several authors8-16 have characterized cowpea accessions using various markers however, morphological and agronomic traits remain valuable to plant breeders16. This is particularly so as identification of key traits of the germplasm enhances the more efficient selection of parents for breeding. Similarly, variation studies in cultivated crops provides valuable information for intraspecific hybridization towards the mapping of traits for the establishment of new populations17.
The need for proper understanding of the variability that exists in cowpea accessions is highly necessary to formulate and accelerate the conventional breeding programme. Collection, evaluation and quantification of diverse variability existing for different characters in cowpea accessions would help to classify cowpea into groups/clusters and identify desirable traits. This would in turn assist breeders in efficient selections. In view of this, the present study aimed to evaluate the extent of variability existing for different morphological characters in ten cowpea accessions collected from ten African countries which will be of immense use for breeders and farmers to choose cowpea accessions of interest for different breeding program.
MATERIALS AND METHODS
Plant materials: All the cowpea seeds from African countries that were used for this study were obtained from the Genetic Resources Unit of the International Institute of Tropical Agriculture (IITA), Ibadan, Oyo State Nigeria. The denomination of the collected accessions was assigned by IITA. The assigned denomination (accession number), cultivar name and country of origin were presented in Table 1.
Experimental site: The study was carried done in the Botanical Garden screen house of the University of Lagos, Nigeria (Latitude 6°30.8800 N: Longitude 3°23.9440 E). The temperature ranged from 34.7°C during the daytime to 28.7°C at night, while maximum and minimum humidity were 75% and 55%, respectively.
Experimental design: Clean plastic bowls filled with loamy soil were used for the planting in an incomplete randomized block experimental set up of five replicates each. Four rows were marked per plot with intra row spacing of 15 cm and inter row spacing of 50 cm. The planting method used was drilling. The spacing was done within the screen house in other to give equal chances of survival to the crop and also avoid overcrowding. The germination period was observed for all the samples and documented. The plants were watered with an equal volumes of water when needed all through the period of the study. The entire screen house and the surroundings were kept clean and all weeds around were handpicked. The experiment was observed from 0 to 12 weeks.
| Table 1: | Cowpea accession used in the study | |||
| Accession number | Cultivar name | Country of origin |
| TVu-144 | SVS-4 | Tanzania |
| TVu-1236 | Early Brown eye (-A) | South Africa |
| TVu-10847 | EX KILALA MARKET | Kenya |
| TVu-11825 | MW80-388 | Malawi |
| TVu-12191 | Ex Tangasia | Ghana |
| TVu-12995 | Ex Beforona | Madagascar |
| TVu-13840 | G-39 | Central African Republic |
| TVu-14316 | B-306 | Botswana |
| TVu-14976 | Matameye-Magaria (Yaouri Village) | Niger Republic |
| TVu-16071 | Afgoi Market Place | Somalia |
Data collection: Cowpea data were collected following the morphological descriptors of the International Board for Plant Genetic Resources18 at different growth stages. Morphological evaluation was done using a modified method of Padulosi and Ng19. A total of thirty-seven characters were assessed comprising both qualitative and quantitative characters of the germinated plants. To avoid being biased, five representatives of each of the ten accessions studied were chosen and observed along the two diagonals of the observation square. On each of the studied accessions, both quantitative and qualitative traits were observed, measured with a meter rule and then coded appropriately for data entry.
Data analysis: Descriptive statistics of the mean and standard error of the data collected were calculated using Microsoft Excel 2013. Pair wise analysis was carried out by computing distance matrices for the characters assessed following Onuminya et al.20. Also, using NTSYS-pc 2.02j software package, a sequential, hierarchical and nested (SAHN) cluster analysis was carried out. Based on Nei genetic distances, dendrogram was generated following Quesada et al.21.
RESULTS
All the five replicates of cowpea accession germinated except Tvu-10847 and Tvu-11825 in which only three and two replicates germinated, respectively (Fig. 1). A large number of the seeds germinated within three days while others germinated after four days. Percentage germination was highest in TVu-13840 (100%) followed by TVu-16071 (93%) while the least percentage germination was observed in TVu-11825 (47%) (Table 2).
There were variations in the quantitative traits evaluated among the studied cowpea accession. The plant height varies amongst the accessions throughout the growth period. At the end of the 12th week, TVu-10847 had the highest average plant height (283.9 cm) followed by TVu-11825 (248.3 cm) while TVu-1236 had the lowest average plant height (137 cm) (Table 3).

|
| Table 2: | Percentage germination of the cowpea accessions studied | |||
| Accession number | Replicate 1 |
Replicate 2 |
Replicate 3 |
Replicate 4 |
Replicate 5 |
Total (15) |
Germination (%) |
| TVu-144 | 3 |
3 |
2 |
2 |
2 |
12 |
80 |
| TVu-1236 | 1 |
1 |
2 |
3 |
3 |
10 |
67 |
| TVu-10847 | 3 |
2 |
2 |
2 |
0 |
9 |
60 |
| TVu-11825 | 3 |
0 |
2 |
2 |
0 |
7 |
47 |
| TVu-12191 | 2 |
3 |
3 |
3 |
1 |
12 |
80 |
| Tvu-12995 | 1 |
2 |
3 |
2 |
3 |
11 |
73 |
| TVu-13840 | 3 |
3 |
3 |
3 |
3 |
15 |
100 |
| TVu-14316 | 1 |
2 |
2 |
3 |
3 |
11 |
73 |
| TVu-14976 | 1 |
3 |
1 |
3 |
2 |
10 |
67 |
| TVu-16071 | 2 |
3 |
3 |
3 |
3 |
14 |
93 |
| Table 3: | Height of the cowpea accessions studied across different weeks | |||
| Accession number | Week 2 |
Week 4 |
Week 6 |
Week 8 |
Week 10 |
Week 12 |
| TVu-144 | 9.6±0.73 |
20.2±0.60 |
59.3±1.11 |
144.0±18.30 |
184.0±15.20 |
192.6±14.70 |
| TVu-1236 | 11.6±0.48 |
20.1±0.62 |
59.5±0.40 |
107.0±7.39 |
127.9±8.63 |
137.0±5.79 |
| TVu-10847 | 8.6±0.50 |
17.0±0.45 |
56.0±0.42 |
200.0±13.70 |
239.9±26.30 |
283.9±32.10 |
| TVu-11825 | 11.0±0.22 |
19.5±1.18 |
58.4±1.06 |
136.0±5.30 |
188.0±18.50 |
248.3±5.77 |
| TVu-12191 | 11.0±0.76 |
20.2±1.63 |
60.2±0.90 |
146.0±8.20 |
140.8±12.50 |
149.7±9.30 |
| Tvu-12995 | 8.7±0.60 |
19.0±0.91 |
58.5±1.11 |
128.0±12.30 |
150.0±17.10 |
149.2±18.40 |
| TVu-13840 | 7.9±0.80 |
14.9±0.37 |
54.6±1.34 |
126.0±17.4 |
159.0±15.40 |
187.3±13.50 |
| Tvu-14316 | 8.9±0.76 |
17.8±1.79 |
56.8±1.21 |
115.0±16.20 |
135.2±19.70 |
165.0±14.10 |
| TVu-14976 | 7.8±0.34 |
15.5±0.16 |
49.7±7.30 |
109.0±21.80 |
138.3±10.70 |
153.2±25.20 |
| TVu-16071 | 11.7±0.25 |
21.9±0.84 |
61.7±0.82 |
113.0±15.10 |
138.3±26.60 |
164.1±21.20 |
| Values represent mean±SE (standard error) | ||||||
Four cowpea accessions (TVu-144, Tvu-10847, Tvu-14976 and Tvu-16071) showed semi-erect growth habit, three (Tvu-1236, Tvu-11825 and Tvu-14316) showed intermediate growth habit, two (Tvu-12191 and Tvu-13840) showed semi-prostrate growth habit while only Tvu-12995 displayed erect growth habit (Table 4).
Almost all the cowpea accession exhibited globose leaf shape with the exception of Tvu-144 and Tvu-12995 which displayed sub-globose and sub-hastate leaf shape respectively (Fig. 2, Table 4). Most of the studied cowpea accession showed a slight tendency to twine although, Tvu-10847 and Tvu-11825 displayed a pronounced twining tendencies (Table 4). The leaf surface area of the studied cowpea varies between accessions. Tvu-1236 (278 cm2) recorded the largest surface area, while Tvu-16071 (70 cm2) had the lowest surface area. The rachis length and petiole length recorded were not within the same range. The Tvu-10847 had the highest mean rachis length (2.83 cm) and mean petiole length (11.73 cm) among studied accessions (Table 4).
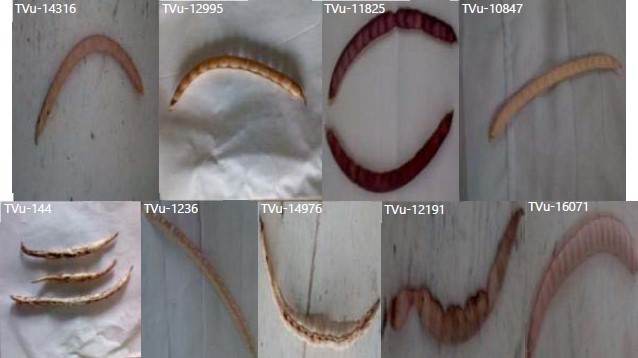
The accession numbers: The Tvu-11825, Tvu-12191 and Tvu-14316 showed extensive pigmentation, while Tvu-144, Tvu-12995, Tvu-14976 and Tvu-16071 showed no pigmentation (Table 5). Only Tvu-1236 and Tvu-12995 showed no pigmented flower while the remaining accessions showed either wing pigmented or completely pigmented flower (Fig. 3, Table 5). Accession Tvu-144 and Tvu-11825 showed splashed and uniform pattern of pigmentation on their pods respectively while the remaining seven accessions has no pigmentation on their (Fig. 4, Table 5).
The raceme position observed among the studied accession indicates that two (TVU-12191 and TVU-14976) accessions had above canopy, four (TVU-1236, TVU-11825, TVU-144 and TVU-14316) accessions had throughout canopy and three (TVU-10847, TVU-12995 and TVU-16071) accessions had upper canopy (Table 5). The flower color was also observed which include violet (n = 6), white (n = 2) and mauve pink (n = 1) among the studied cowpea accession (Fig. 3).

|

|
Other qualitative characters evaluated among the studied accession were presented in Table 5. All the cowpea accession developed straight pod shape except Tvu-14316 whose pod shape was slightly curved (Fig. 4). Variation in seed shape was also observed, accession (TVU-144, TVU-11825, TVU-12191 and TVU-12995) developed kidney-shaped seeds, TVU-1236 and TVU-14316 had globose-shaped seeds, TVU-14976 and TVU-16071 showed rhomboid-shaped seeds, while TVU-10847 showed ovoid-shaped seeds (Fig. 5, Table 5). Among the ten studied accession, only Tvu-13840 neither developed to flowering nor developed pods. Therefore, pod characters, seed characters and floral characters were not evaluated for the aforementioned accession.
Days of first flowering observed among the accessions studied range between 54 days in TVu-1236 to 95 days in TVu-11825 and Tvu-16071, while days of first pod range between 67 days in TVu-1236 to 114 days in TVu-11825. Peduncle length ranges from 1.82 cm in TVu-12191 which was the lowest to 15.5 cm in TVu-11825 which is the highest. Pod length was lowest in TVu-12995 (8.63 cm) and highest in TVu-10847 (18.83 cm). The TVu-14976 (0.77 g) recorded the lowest pod weight when evaluated, while TVu-11825 (5.20 g) recorded the highest. The seed weight also varies and range between 726-2381 mg per ten seed. Other quantitative characters evaluate among the studied cowpea accession were presented in Table 6.
|

|
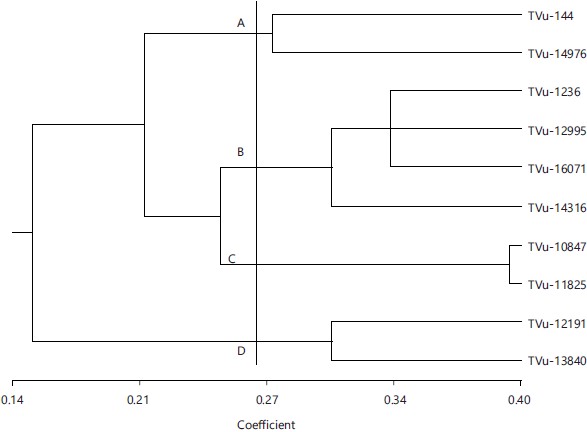
Cluster analysis of morphological characteristics: The cluster analysis (Fig. 6) resolved the morphological characters of ten sampled accession at about 0.40 similar coefficient while the accessions were grouped as one at similarity level of 0.14. There are various stages between the two extremes which led to different clusters, with the accession survey as the Operational Taxonomic Unit (OTU). As the similarity index decreases, the number of clusters is reduced and gave rise to only one cluster of 0.153 levels. At similarity coefficient of 0.27, a truncated line (phenol line) demarcated the UPGMA dendrogram into four clusters namely A, B, C and D.
| Table 4: | Vegetative morphological characters of the ten cowpea accessions studied | |||
| Accession number | LC |
PV |
GP |
LS |
GH |
ECC |
TT |
NMSN |
NMB |
MRL |
MPL |
MPH |
MTLL |
MTLW |
LSA |
|
| TVu-144 | G |
V |
Det |
SG |
SE |
B30 |
Sl |
9 |
12 |
2.34 |
10.14 |
59.26 |
9.3 |
5.76 |
96 |
|
| TVu-1236 | DG |
V |
Det |
Gl |
Int |
T10 |
Sl |
7 |
8 |
2.44 |
10.56 |
59.48 |
10 |
6.48 |
278 |
|
| TVu-10847 | G |
Int |
Det |
Gl |
SE |
T41 |
Pr |
9 |
11 |
2.83 |
11.73 |
56.03 |
10.8 |
7.53 |
147 |
|
| TVu-11825 | G |
Int |
Det |
Gl |
Int |
T41 |
Pr |
7 |
17 |
2.43 |
10.23 |
58.43 |
9.93 |
7.73 |
161 |
|
| TVu-12191 | PG |
NV |
Indet |
Gl |
SP |
B30 |
Int |
10 |
9 |
2 |
7.2 |
60.16 |
6.83 |
5.02 |
103 |
|
| TVu-12995 | G |
Int |
Det |
SH |
Er |
T60 |
Sl |
8 |
10 |
2.1 |
9.3 |
58.46 |
11.43 |
6.48 |
128 |
|
| TVu-13840 | PG |
NV |
Indet |
Gl |
SP |
- |
Int |
8 |
17 |
1.56 |
4.1 |
54.64 |
8.47 |
4.46 |
112 |
|
| TVu-14316 | G |
Int |
Det |
Gl |
Int |
M60 |
Sl |
8 |
9 |
1.64 |
9 |
55.86 |
9.37 |
6.32 |
118 |
|
| TVu-14976 | DG |
NV |
Indet |
Gl |
SE |
B30 |
Sl |
9 |
10 |
1.78 |
6.68 |
49.66 |
9 |
4.82 |
111 |
|
| TVu-16071 | DG |
Int |
Det |
Gl |
SE |
R40 |
Sl |
8 |
13 |
1.78 |
9 |
61.68 |
8.3 |
5.9 |
70 |
|
| LC: Leaf color: G: Green, PG: Pale green, DG: Dark green, PV: Plant vigor: V: Vigorous, NV: Non-vigorous, Int: Intermediate, GP: Growth pattern: Det: Determinate, Indet: Indeterminate, LS: Leaf shape: Gl: Globose, SG: Sub globose, Ha: Hastate, SH: Sub hastate, GH: Growth habit: SE: Semi erect, Er: Erect, SP: Semi prostrate, Int: Intermediate, ECC: Eye color code, R40: Red, B30: Solid black, T60: Brown, M60: Solid brown over Tan, T10: Tan, T41: Light orange tan, TT: Twining tendency: Sl: Slight, Pr: Pronounced, Int: Intermediate, NMSN: Number of main stem node, NMB: Number of main branches, MRL: Mean rachis length (cm), MPL: Mean petiole length (cm), MPH: Mean plant height (cm), MTLL: Mean terminal leaflet length (cm), MTLW: Mean terminal leaflet width (cm) and LSA: Leaf surface area (cm2) | ||||||||||||||||
| Table 5: | Qualitative reproductive characters of the ten African accessions of cowpea studied | |||
| Accession number | PP |
PPO |
PAP |
RP |
PF |
Seed shape |
TT |
Pod shape |
Flower color |
SCP |
Seed color |
| TVu-144 | None |
Pigmented |
Erect |
TC |
CP |
Kidney |
Rough |
Straight |
Violet |
SC |
DBBS |
| TVu-1236 | Slight |
NP |
30-90o |
TC |
NP |
Globose |
Smooth |
Straight |
White |
NC |
Cream |
| TVu-10847 | Moderate |
NP |
Pendant |
UC |
WP |
Ovoid |
Rough |
Straight |
Violet |
NC |
LB |
| TVu-11825 | Extensive |
Pigmented |
30-90o |
TC |
WP |
Kidney |
Rough |
Straight |
Violet |
SC |
LB |
| TVu-12191 | Extensive |
NP |
Erect |
AC |
WP |
Kidney |
Smooth |
Straight |
Violet |
C |
DBBS |
| TVu-12995 | None |
NP |
Pendant |
UC |
NP |
Kidney |
Smooth |
Straight |
White |
NC |
BWP |
| TVu-13840 | Extensive |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
| TVu-14316 | Extensive |
NP |
30-90o |
WP |
Globose |
Smooth |
Curved |
Violet |
NC |
LCDP |
|
| TVu-14976 | None |
NP |
30-90o |
AC |
WP |
Rhomboid |
Rough |
Straight |
Mauve Pink |
SC |
DCBP |
| TVu-16071 | None |
NP |
Pendant |
UC |
WP |
Rhomboid |
Smooth |
Straight |
Violet |
NC |
Pale Red |
| PP: Pigmentation on plant, PPO: Pigmentation on Pod, NP: Not pigmented, PAP: Pod attachment to peduncle, RP: Raceme position, TC: Throughout canopy, UC: Upper canopy and AC: Above canopy, PF: Pigmentation on flower, WP: Wing pigmented, CP: Completely pigmented and NP: Not pigmented, TT: Testa texture, SCP: Seed crowding in pod, C: Crowded, SC: Semi crowded, NC: Not crowded, Seed color BWP: Brown (White patches) and DBBS: Dark brown | |||||||||||
|
Cluster A: It consist of two accessions (TVu-144 and TVu-14976) which were closely related at about 0.272 similarity coefficient. This closeness can be attributed to the fact that these two accessions have the same pattern of pigmentation on plant, plant growth habit, twining tendency, testa texture, pod shape, eye color, seed crowding in pod, number of main stem nodes and days to first ripe pod.
Cluster B: It consist of four accessions (TVu-1236, TVu-12995, TVu-16071 and TVu-14316) while three of these accessions indicates a polytomy which could suggest that the knowledge of their similarity is not clear. The TVu-1236 and TVu-16071 were closely related at about 0.34 similarity coefficient. This closeness can be linked to the fact that these two accessions have the same pattern of pigmentation on green pod, leaf shape, twining tendency, testa texture, pod shape, leaf color, seed crowding in pod, plant growth pattern and seed width. The TVu-12995 and TVu-14316 were closely related at about 0.310 similarity coefficient. This closeness can be traced to the fact that the two accessions have the same pattern of pigmentation on green pod, twining tendency, testa texture, leaf color, plant vigor, seed crowding in pod, plant growth pattern, number of main stem nodes and days to first flowering.
Cluster C: It consists of two accessions (TVu-10847 and TVu-11825) which were closely related at about 0.398 similarity coefficient. This closeness can be attributed to the fact that these two accessions have the same leaf shape, twining tendency, eye color, testa texture, pod shape, pattern of pigmentation on flower, leaf color, plant vigor, seed crowding in pod, plant growth pattern, flower color and seed color.
| Table 6: | Quantitative reproductive characters of the ten African accessions of cowpea studied | |||
| Accession number | DFF |
DFRP |
PL (mm) |
PW (cm) |
POL (cm) |
NPP |
NLP |
NPPP |
Seed length (cm) |
Seed width (cm) |
Seed weight (mg/10 seed) |
| TVu-144 | 60 |
81 |
2.37±0.57 |
1.00±0.30 |
12.80±0.72 |
1 |
12 |
4 |
0.60±0.10 |
0.44±0.06 |
790 |
| TVu-1236 | 54 |
67 |
2.22±0.40 |
1.62±0.76 |
10.72±1.52 |
1 |
11 |
3 |
0.66±0.06 |
0.48±0.05 |
879 |
| TVu-10847 | 85 |
108 |
13.70±1.49 |
4.00±1.93 |
18.83±2.97 |
2 |
14 |
3 |
0.92±0.05 |
0.84±0.05 |
2381 |
| TVu-11825 | 95 |
114 |
15.50±0.00 |
5.20±0.00 |
17.7±0.00 |
1 |
14 |
1 |
0.84±0.06 |
0.62±0.05 |
1267 |
| TVu-12191 | 59 |
72 |
1.82±0.30 |
0.86±0.17 |
11.7± 0.83 |
1 |
9 |
2 |
0.68±0.05 |
0.52±0.05 |
726 |
| TVu-12995 | 59 |
77 |
3.48±0.71 |
0.80±0.22 |
8.63±1.84 |
2 |
5 |
4 |
0.82±0.08 |
0.54±0.06 |
1508 |
| TVu-13840 | - |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
| TVu-14316 | 59 |
74 |
2.50±0.52 |
1.70±0.86 |
11.92±2.02 |
1 |
10 |
3 |
0.76±0.06 |
0.52±0.05 |
1699 |
| TVu-14976 | 66 |
81 |
1.87±0.15 |
0.77±0.06 |
10.70±1.25 |
2 |
6 |
3 |
0.78±0.13 |
0.56±0.09 |
1345 |
| TVu-16071 | 95 |
113 |
9.50±0.71 |
1.15±0.49 |
11.95±0.35 |
2 |
9 |
2 |
0.64±0.06 |
0.48±0.05 |
1005 |
| DFF: Days to first flowering, DFRP: Days to first ripe pod, PL: Peduncle length, PW: Pod weight, POL: Pod length, NLP: Number of locules per pod, NPP: Number of pod per peduncle and NPPP: Number of pod per plant | |||||||||||
Cluster D: It consists of two accessions (TVu-12191 and TVu-13840) which were closely related at about 0.310 similarity coefficient. This closeness can be linked to the fact that the two accessions have the same pattern of pigmentation on plant, pattern of pigmentation on green pod, leaf shape, plant growth habit, leaf color, plant vigor and plant growth pattern.
DISCUSSION
Previous studies on cowpea indicate that morphological traits influences the potential yield of cowpea8-10,22. Also, a number of morphological characters were found to be of great importance to assess the level of genetic variability and have led to a better classification of cowpea species18,23-27. This study compared ten cowpea genotypes originating from various countries in Africa at morphological level. Both quantitative and qualitative traits were evaluated. The wide range of data obtained from majority of the traits suggested that genetic variability exists in the accessions hence they can be improved through plant breeding programmes. High variation was observed in the quantitative characters evaluated in the cowpea genotype while a few qualitative traits such as leaf shape, pod shape and pattern of pigmentation on pod showed less variation. The influence of these traits on cowpea contributes directly or indirectly to the variation that exist among these cultivars. The high variation observed in peduncle length, pod weight, pod length, number of locules per pod, number of pod per peduncle and number of pod per plant could be linked to genetic factors including those control time-factor for assimilates. These variations are of prime importance to framers, consumers and plant breeders, e.g. peduncle length of cowpea helps to determine pods position on the plant, which would in turn ease the visibility of pods on the crop canopy. Thereby, becoming an important character with respect to harvesting28. The study of Doumbia et al.29 also reported that pod length and number of locules/pods has effects on seed yield and these traits can also be genetically controlled.
The weights of ten seeds of the ten accessions varies between 726-2381 mg, this finding also corroborate the values obtained for seed weight by Gbaguidi et al.30 which was found to be between 400 mg and 2375 mg. The observed flower color of the accessions evaluated in this study showed that 66.7% of the genotypes developed violet flowers, 22.2% white flowers and 11.1% developed mauve-pink flowers which is in agreement with the observation of Yasin et al.28. Leaf color intensity also varied from pale to dark green. The study showed 50% of the accession having green leaves while dark green and pale green leaf colors were found in 30% and 20%, respectively of the cowpea genotypes. Also, 77.8% of the accession did not develop pod pigmentation while 22.8% had pigmentation on their pod. The variation observed in the intensity of green coloration on leaves, flower pigmentation, leaf shape, seed color, seed eye color and twinning tendency highlighted the presence of a diversity of morphological characters between the cowpea accessions. The results of this study was also consistent with the reports of Amusa et al.31.
Majority of the genotypes displayed globose leaf shape making up 80% of the cowpea accession while sub-hastate and sub-globose were each found in 10% of the genotypes. The kidney, ovoid, globose and rhomboid seed shape was observed in this study. Furthermore, variation in coefficient of diversity was recorded for various stages of the plant development, this is in agreement with the results reported by Amusa et al.31. In some genotypes (TVu-1236) early flowering was observed (54 days after germination) while in others (TVu-11825 and TVu-16071) flowering was late (95 days after germination). According to Omomowo and Babalola32 short flowering period is an advantage at high temperatures and this also helps in avoiding low air humidity.
The dendrogram resulting from the hierarchical clustering of the quantitative and qualitative characters revealed four major clusters (A, B, C and D). This dendrogram showed a perfect correspondence with the morphological classification of the accessions studied using the quantitative and qualitative characters. This indicate that the accessions are not similar genetically since there are some clear distinctions among the accessions collected from the ten African countries. Furthermore, the use of molecular method to elucidate and support the morphological evidences established in this study is also of prime importance.
CONCLUSION
Genetic diversity based on morphological characters are affected by variations in environmental conditions as well as limited number of morphological markers thus accounting for the relatively low similarity index recorded amongst the accessions studied. However, significant variations in several morphological traits were detected. Also, the late flowering and date to maturity of some of the studied cowpea accession suggest that a number of these genotypes are photoperiod sensitive.
SIGNIFICANCE STATEMENT
Owing to the need for proper understanding of the variability that exist in cowpea accessions to formulate and accelerate conventional breeding program, this study aimed to evaluate the extent of variability existing for different morphological characters in ten cowpea accessions collected from ten African countries. Both quantitative and qualitative vegetative and reproductive characters of the plant were assessed. Results showed great diversity in both characters which assisted in classifying the accessions into four clusters. The variations recorded would assist breeders in efficient selections for their breeding program.
ACKNOWLEDGMENT
Special thanks to Mr. Mubarak Ogunkanmi for the technical support provided during the planting experiments.
REFERENCES
- Maxted, N., P. Mabuza-Diamini, H. Moss, S. Padulosi, A. Jarvis and L. Guarino, 2004. Systematic and Ecogeographic Studies on Crop Genepools 11: An Ecogeographic Study African Vigna. Bioversity International, Rome, Italy, ISBN-13: 978-92-9043-637-9, Pages: 454.
- Pasquet, R.S., 1996. Wild Cowpea (Vigna unguiculata) Evolution. In: Advances in Legume Systematics Part 8: Legumes of Economic Importance, Pickersgill, B. (Ed.), Royal Botanic Gardens, Kew, Richmond, United Kingdom, ISBN: ISBN: 9781842465615, pp: 95-100.
- Simion, T., 2018. Breeding cowpea Vignaunguiculata L. Walp for quality traits. Ann. Rev. Res., 3: 45-51.
- Goufo, P., J.M. Moutinho-Pereira, T.F. Jorge, C.M. Correia and M.R. Oliveira et al., 2017. Cowpea (Vigna unguiculata L. Walp.) metabolomics: Osmoprotection as a physiological strategy for drought stress resistance and improved yield. Front. Plant Sci., Vol. 8.
- Ahenkora, K., H.K.A. Dapaah and A. Agyemang, 1998. Selected nutritional components and sensory attributes of cowpea (Vigna unguiculata [L.] Walp) leaves. Plant Foods Hum. Nutr., 52: 221-229.
- Nielsen, S.S., T.A. Ohler and C.A. Mitchell, 1997. Cowpea Leaves for Human Consumption: Production, Utilization and Nutrient Composition. In: Advances in Cowpea Research, Singh, B.B., D.R.M. Raj, K.E. Dashiell and L.E.N. Jackai (Eds.), IITA, Ibadan, Nigeria, ISBN: 9789781311109, pp: 326-332.
- Fatokun, A.C., 2002. Breeding Cowpea for Resistance to Insects Pests, Attempted Crosses Between Cowpea and Vigna vexillata. In: Challenges and Opportunities for Enhancing Sustainable Cowpea Production, Fatokun, C.A., S.A. Tarawali, B.B. Singh, P.M. Kormawa and M. Tamo (Eds.), IITA, Ibadan, Nigeria, ISBN: 9789781311901, pp: 52-61.
- Gerrano, A.S., Z.G. Thungo and S. Mavengahama, 2022. Phenotypic description of elite cowpea (Vigna ungiculata L. Walp) genotypes grown in drought-prone environments using agronomic traits. Heliyon, Vol. 8.
- Mafakheri, K., M.R. Bihamta and A.R. Abbasi, 2017. Assessment of genetic diversity in cowpea (Vigna unguiculata L.) germplasm using morphological and molecular characterisation. Cogent Food Agric., Vol. 3.
- Ogunkanmi, A., O. Ogundipe, L. Omoigui, A. Odeseye and C. Fatokun, 2019. Morphological and SSR marker characterization of wild and cultivated cowpeas (Vigna unguiculata L. Walp). J. Agric. Sci., 64: 367-380.
- Ogunkanmi, L.A., O.T. Ogundipe and C.A. Fatokun, 2014. Molecular characterization of cultivated cowpea (Vigna unguiculata L. Walp) using simple sequence repeats markers. Afr. J. Biotechnol., 13: 3464-3472.
- Omoigui, L.O., M. Yeye, B. Ousmane, B. Gowda and M.P. Timko, 2012. Molecular characterization of cowpea breeding lines for striga resistance using SCAR markers. J. Agric. Sci. Technol. B, 2: 362-367.
- Tanhuanpaa, P. and O. Manninen, 2012. High SSR diversity but little differentiation between accessions of Nordic timothy (Phleum pretense L.). Hereditas, 149: 114-127.
- Odeigah, P.G.C. and A.O. Osanyinpeju, 1996. Seed protein electrophoretic characterization of cowpea (Vigna unguiculata) germplasm from IITA gene bank. Genet. Resour. Crop Evol., 43: 485-491.
- Alghamdi, S.S., M.A. Khan, H.M. Migdadi, E.H. El-Harty, M. Afzal and M. Farooq, 2019. Biochemical and molecular characterization of cowpea landraces using seed storage proteins and SRAP marker patterns. Saudi J. Biol. Sci., 26: 74-82.
- Krichen, L., J.M. Audergon and N. Trifi-Farah, 2012. Relative efficiency of morphological characters and molecular markers in the establishment of an apricot core collection. Hereditas, 149: 163-172.
- Animasaun, D.A., S. Oyedeji, O.T. Mustapha and M.A. Azeez, 2015. Genetic variability study among ten cultivars of cowpea (Vigna unguiculata L. Walp) using morpho-agronomic traits and nutritional composition. J. Agric. Sci. Sri Lanka, 10: 119-130.
- Nkhoma, N., H. Shimelis, M.D. Laing, A. Shayanowako and I. Mathew, 2020. Assessing the genetic diversity of cowpea [Vigna unguiculata (L.) Walp.] germplasm collections using phenotypic traits and SNP markers. BMC Genet., Vol. 21.
- Podulosi, S. and N.Q. Ng, 1993. A useful and unexploited herb, Vigna marina (Leguminosae-Papilionoideae) and the taxonomic revision of its genetic diversity. Bull. Jard. Bot. Nat. Belg., 62: 119-126.
- Onuminya, T.O., A.A. Kalesanwo and O.T. Ogundipe, 2021. Taxonomic studies of the genus Placodiscus (Sapindaceae: Lepisantheae) in West Africa and Cameroon. Phytologia Balcan., 27: 209-220.
- Quesada, E., A. Ventosa, F. Rodriguez-Valera, L. Megias and A. Ramos-Cormenzana, 2009. Numerical taxonomy of moderately halophilic gram-negative bacteria from hypersaline soils. Microbiology, 129: 2649-2657.
- Siise, A. and F.J. Massawe, 2013. Microsatellites based marker molecular analysis of Ghanaian Bambara groundnut (Vigna subterranea (L.) Verdc.) landraces alongside morphological characterization. Genet. Resour. Crop Evol., 60: 777-787.
- Odeseye, A.O., I.F. Ijagbone, S.A. Aladele, H.Y. Gbadegesin and D.J. Nwosu et al., 2022. Characteristics and molecular identification of differentially expressed genes in some cowpea [Vigna unguiculata (L.) Walp.] accessions. J. Agric. Food Res., Vol. 10.
- Brantley, B.B. and C.W. Kuhn, 1983. A genetic abnormality causing virus-like symptoms and sterility in cowpea. Am. Soc. Hortic. Sci., 18: 458-459.
- Fawole, I., 1988. A nonpetiolate leaf mutant in cowpea, Vigna unguiculata (L.) Walp. J. Heredity, 79: 484-487.
- Fery, R.L. and P.D. Dukes, 1994. Genetic analysis of the green cotyledon trait in Southernpea [Vigna unguiculata (L.) Walp.]. J. Am. Soc. Hortic. Sci., 119: 1054-1056.
- Karkannavar, J.C., R. Venugopal and J.V. Goud, 1991. Inheritance and linkage studies in cowpea (Vigna unguiculata) (L.) Walp.). Indian J. Genet. Plant Breed., 51: 203-207.
- Yasin, G., U. Elias, W. Walelign and M. Hussein, 2021. Assessment of genetic diversity in cowpea (Vigna unguiculata) genotypes in Southern Ethiopia based on morpho-agronomic traits. Int. J. Agric. Technol., 17: 1631-1650.
- Doumbia, I.Z., R. Akromah and J.Y. Asibuo, 2013. Comparative study of cowpea germplasms diversity from Ghana and Mali using morphological characteristics. J. Plant Breed. Genet., 1: 139-147.
- Gbaguidi, A.A., P. Assogba, M. Dansi, H. Yedomonhan and A. Dansi, 2015. Agromorphological characterization of cowpea varieties grown in Benin. Int. J. Biol. Chem. Sci., 9: 1050-1066.
- Amusa, O.D., A.L. Ogunkanmi, J.A. Adetumbi, S.T. Akinyosoye and O.T. Ogundipe, 2019. Morpho-genetic variability in F2 progeny cowpea genotypes tolerant to Bruchid (Callosobruchus maculatus). J. Agric. Sci. Belgrade., 64: 53-68.
- Omomowo, O.I. and O.O. Babalola, 2021. Constraints and prospects of improving cowpea productivity to ensure food, nutritional security and environmental sustainability. Front. Plant Sci., 12: 751731.
How to Cite this paper?
APA-7 Style
Onuminya,
T.O., Oyewole,
L., Osundinakin,
M.I., Ogunkanmi,
L.A. (2023). Morphological Characterization of Some Cowpea Accessions from Africa. Trends in Agricultural Sciences, 2(1), 54-66. https://doi.org/10.17311/tas.2023.54.66
ACS Style
Onuminya,
T.O.; Oyewole,
L.; Osundinakin,
M.I.; Ogunkanmi,
L.A. Morphological Characterization of Some Cowpea Accessions from Africa. Trends Agric. Sci 2023, 2, 54-66. https://doi.org/10.17311/tas.2023.54.66
AMA Style
Onuminya
TO, Oyewole
L, Osundinakin
MI, Ogunkanmi
LA. Morphological Characterization of Some Cowpea Accessions from Africa. Trends in Agricultural Sciences. 2023; 2(1): 54-66. https://doi.org/10.17311/tas.2023.54.66
Chicago/Turabian Style
Onuminya, Temitope, Olabisi, Lawal Oyewole, Michael Idowu Osundinakin, and Liasu Adebayo Ogunkanmi.
2023. "Morphological Characterization of Some Cowpea Accessions from Africa" Trends in Agricultural Sciences 2, no. 1: 54-66. https://doi.org/10.17311/tas.2023.54.66

This work is licensed under a Creative Commons Attribution 4.0 International License.




